This vignette is a detailed walk-through of an exploratory
debugging workflow with {ggtrace}, using a Github issue
opened on the {ggxmean}
package by Gina Reynolds. We
define “exploratory debugging” here as the process of learning and
developing simultaneously, assuming little prior knowledge about ggplot
internals.
Setup
If you would like to follow along, you can install the version of the
{ggxmean} package containing the bug that this vignette will address. We
use devtools::dev_mode() here to install and inspect this
version of {ggxmean} in development mode (except I skip installation
here because I already have that version in my dev directory).
library(devtools)
#> Loading required package: usethis
# Current installation version
devtools::package_info("ggxmean", FALSE, FALSE)$source
#> [1] "local"
# Activate development mode
devtools::dev_mode(on = TRUE, path = getOption("devtools.path"))
#> ✔ Dev mode: ON
# devtools::install_github("EvaMaeRey/ggxmean@cbb909c")
# Prior version installed for dev mode
devtools::package_info("ggxmean", FALSE, FALSE)$source
#> [1] "Github (EvaMaeRey/ggxmean@cbb909cee7d4f0a4395722020a75330641e5f54c)"
library(ggxmean)I’ve also attached the code relevant to the issue at hand here, if you would like to familiarize yourself with it before we begin (though we do not assume this as prior knowledge here).
Relevant code from {ggxmean}
# https://github.com/EvaMaeRey/ggxmean/blob/cbb909cee7d4f0a4395722020a75330641e5f54c/R/geom_x_line.R
GeomXline <- ggplot2::ggproto("GeomXline", ggplot2::Geom,
draw_panel = function(data, panel_params, coord) {
ranges <- coord$backtransform_range(panel_params)
data$x <- data$x
data$xend <- data$x
data$y <- ranges$y[1]
data$yend <- ranges$y[2]
GeomSegment$draw_panel(unique(data), panel_params, coord)
},
default_aes = ggplot2::aes(colour = "black", size = 0.5,
linetype = 1, alpha = NA),
required_aes = "x",
draw_key = ggplot2::draw_key_vline
)
# https://github.com/EvaMaeRey/ggxmean/blob/cbb909cee7d4f0a4395722020a75330641e5f54c/R/geom_x_mean.R
StatXmean <- ggplot2::ggproto("StatXmean",
ggplot2::Stat,
compute_group = function(data, scales) {
data.frame(x = mean(data$x))
},
required_aes = c("x")
)
geom_x_mean <- function(mapping = NULL, data = NULL,
position = "identity", na.rm = FALSE, show.legend = NA,
inherit.aes = TRUE, ...) {
ggplot2::layer(
stat = StatXmean, geom = GeomXline, data = data, mapping = mapping,
position = position, show.legend = show.legend, inherit.aes = inherit.aes,
params = list(na.rm = na.rm, ...)
)
}Introduction of the issue
{ggxmean} is a {ggplot2} extension package that offers many features,
including the layer geom_x_mean() which can be used to draw
a line at the mean value of the variable represented by the x-axis.
ggplot(mpg, aes(x = hwy)) +
geom_histogram() +
geom_x_mean()
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.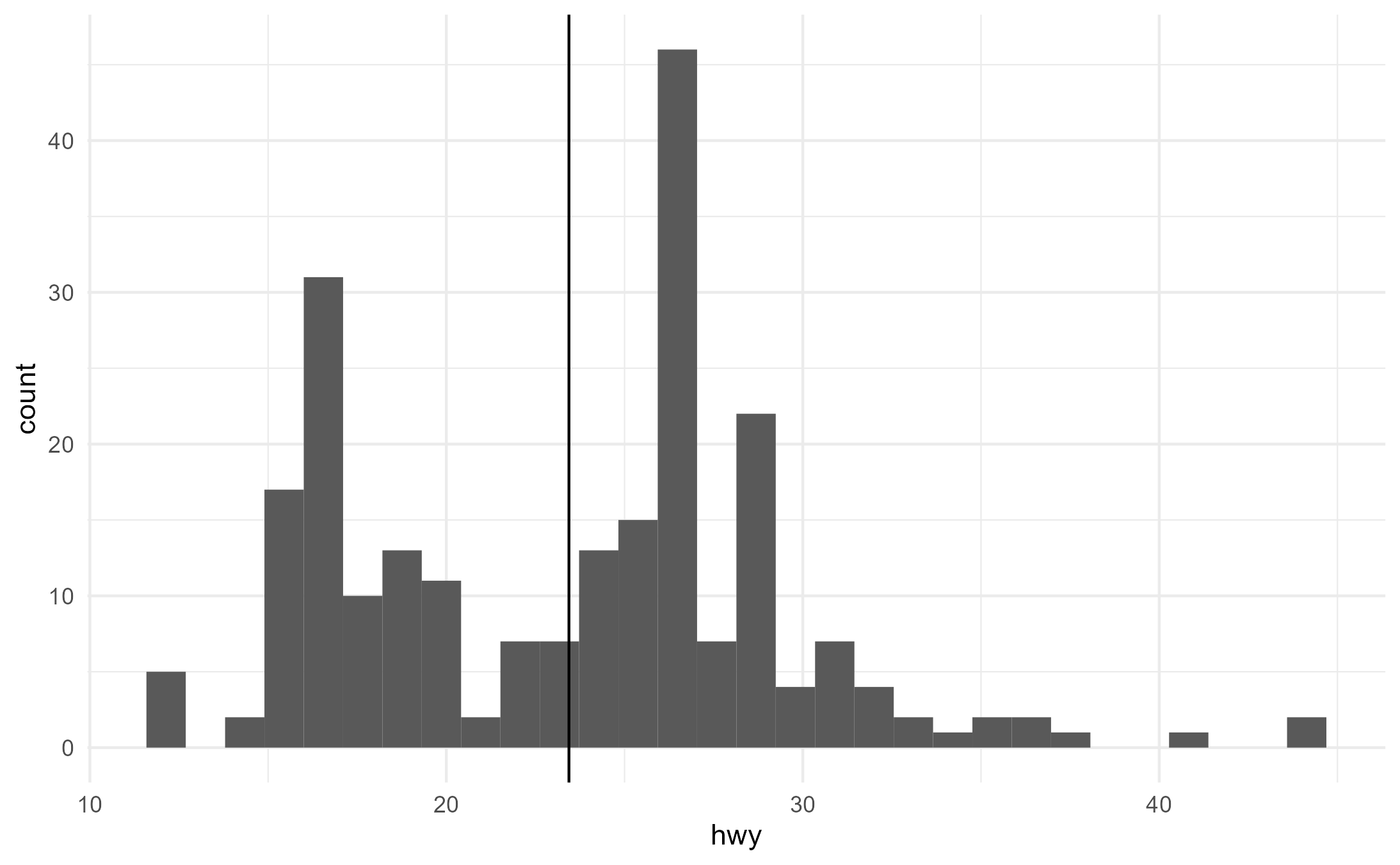
geom_x_mean() works smoothly with groups and facets, as
we might expect from our experience with ggplot.
ggplot(mpg, aes(x = hwy, group = drv)) +
geom_histogram() +
geom_x_mean() +
facet_wrap(~drv)
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.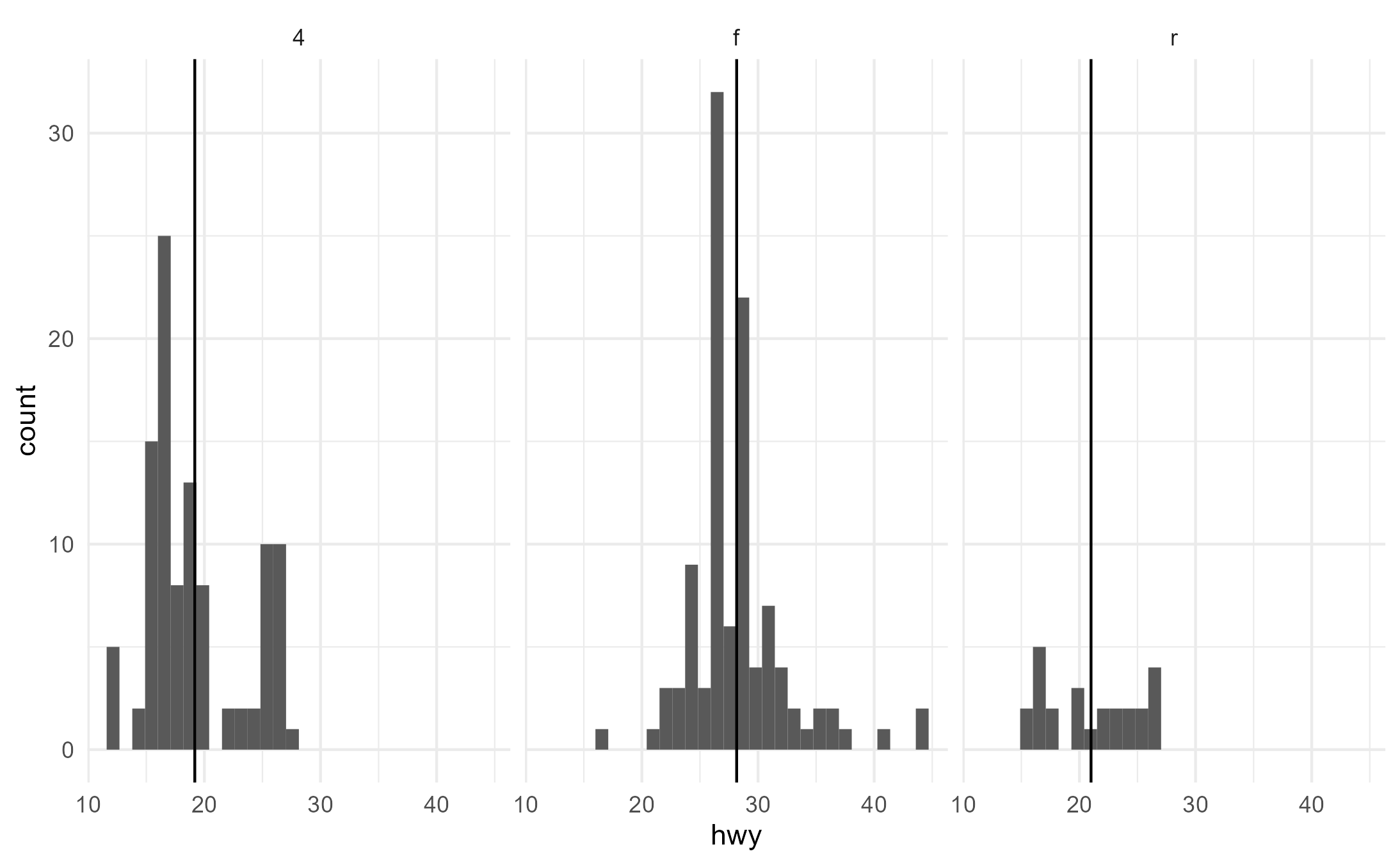
However, something somewhat unexpected happens when we pass map a
discrete variable to an aesthetic that is not understandable by
geom_x_mean(). Despite the fact that the lines do not
understand the fill aesthetic, the aesthetic mapping of
fill = drv nevertheless highjacks the calculation
of grouping for geom_x_mean().
ggplot(mpg, aes(x = hwy, fill = drv)) +
geom_histogram() +
geom_x_mean()
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.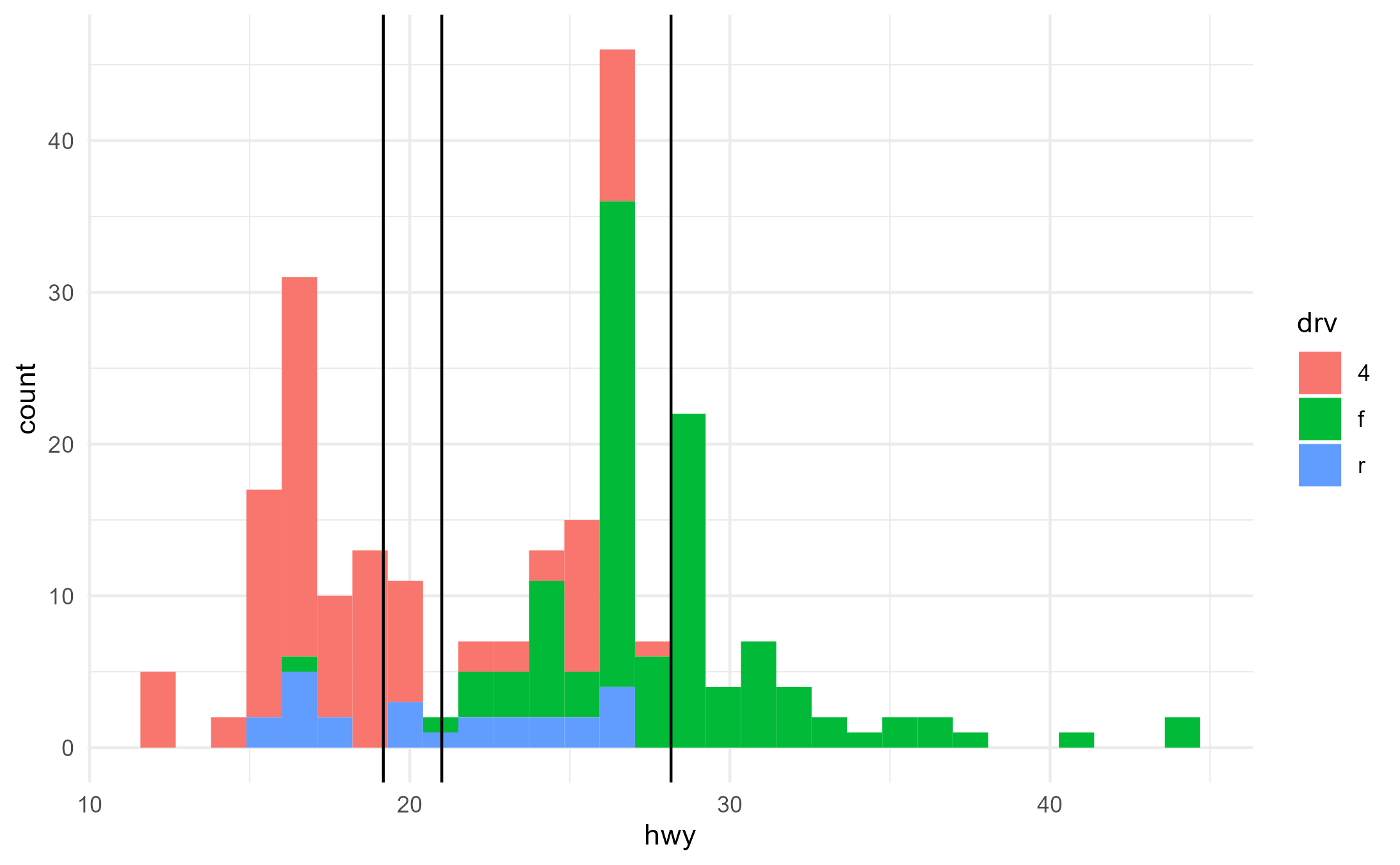
Making a reprex
To isolate the problem, let’s remove geom_histogram()
from the examples.
Here is a minimal reprex (reproducible example) of the issue:
ggplot(mpg) +
aes(x = hwy, fill = drv) +
geom_x_mean()
We can pinpoint and formalize the issue with the help of
layer_data(). We see that in both our expected
group = drv and unexpected fill = drv cases,
we end up with three unique groups (lines) as indicated by the values in
the group column.
p <- ggplot(mpg) +
aes(x = hwy) +
geom_x_mean()
layer_data(p + aes(group = drv))
#> x group PANEL colour size linetype alpha
#> 1 19.17476 1 1 black 0.5 1 NA
#> 2 28.16038 2 1 black 0.5 1 NA
#> 3 21.00000 3 1 black 0.5 1 NA
layer_data(p + aes(fill = drv))
#> fill x PANEL group colour size linetype alpha
#> 1 #F8766D 19.17476 1 1 black 0.5 1 NA
#> 2 #00BA38 28.16038 1 2 black 0.5 1 NA
#> 3 #619CFF 21.00000 1 3 black 0.5 1 NAExpected cases
Here, we correctly get groups for each category of drv
because lines understand the color aesthetic.
p + aes(color = drv)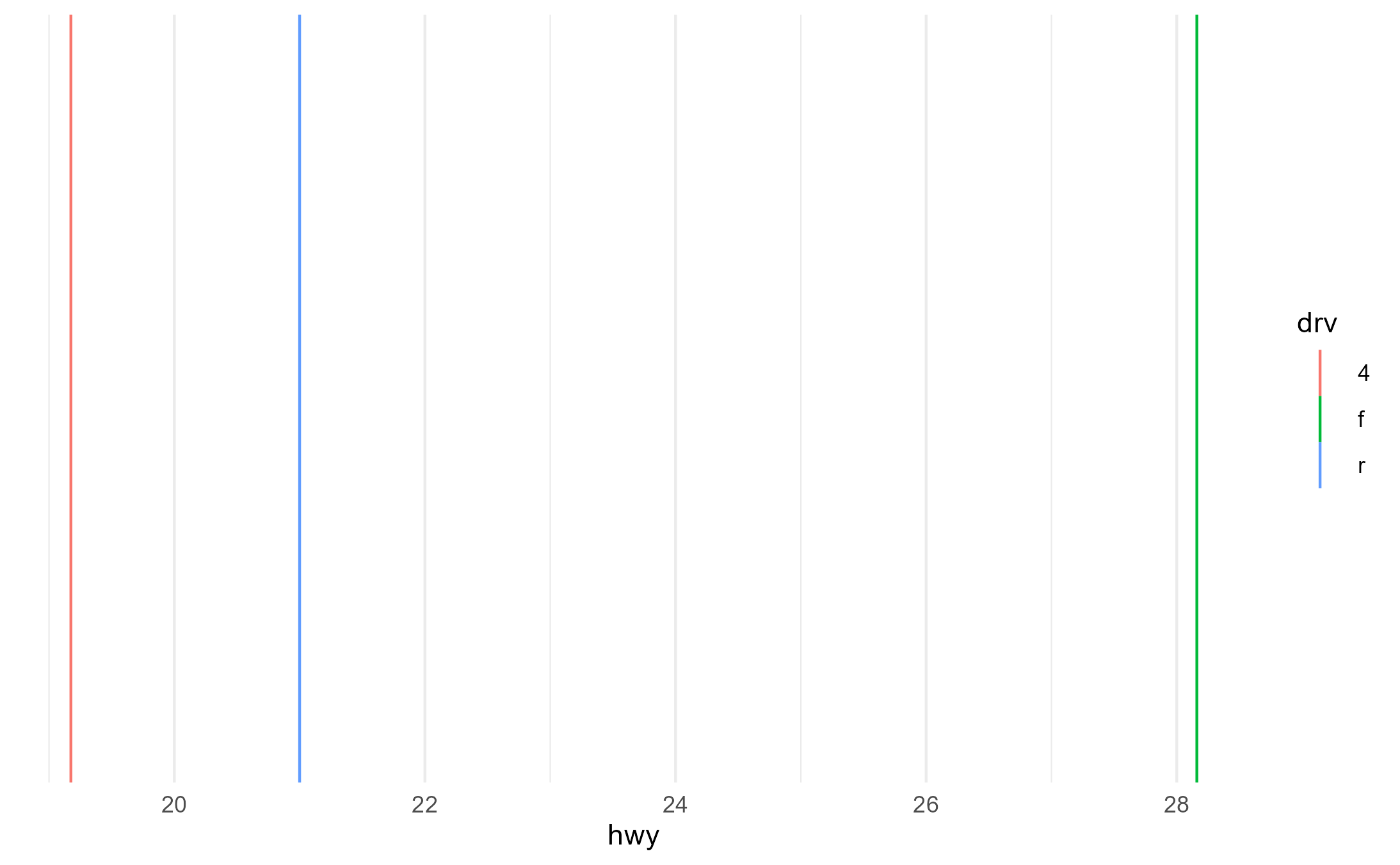
layer_data(p + aes(color = drv))$group
#> [1] 1 2 3We also correctly get groups for each category of drv
when we set the group aesthetic explicitly.
p + aes(group = drv)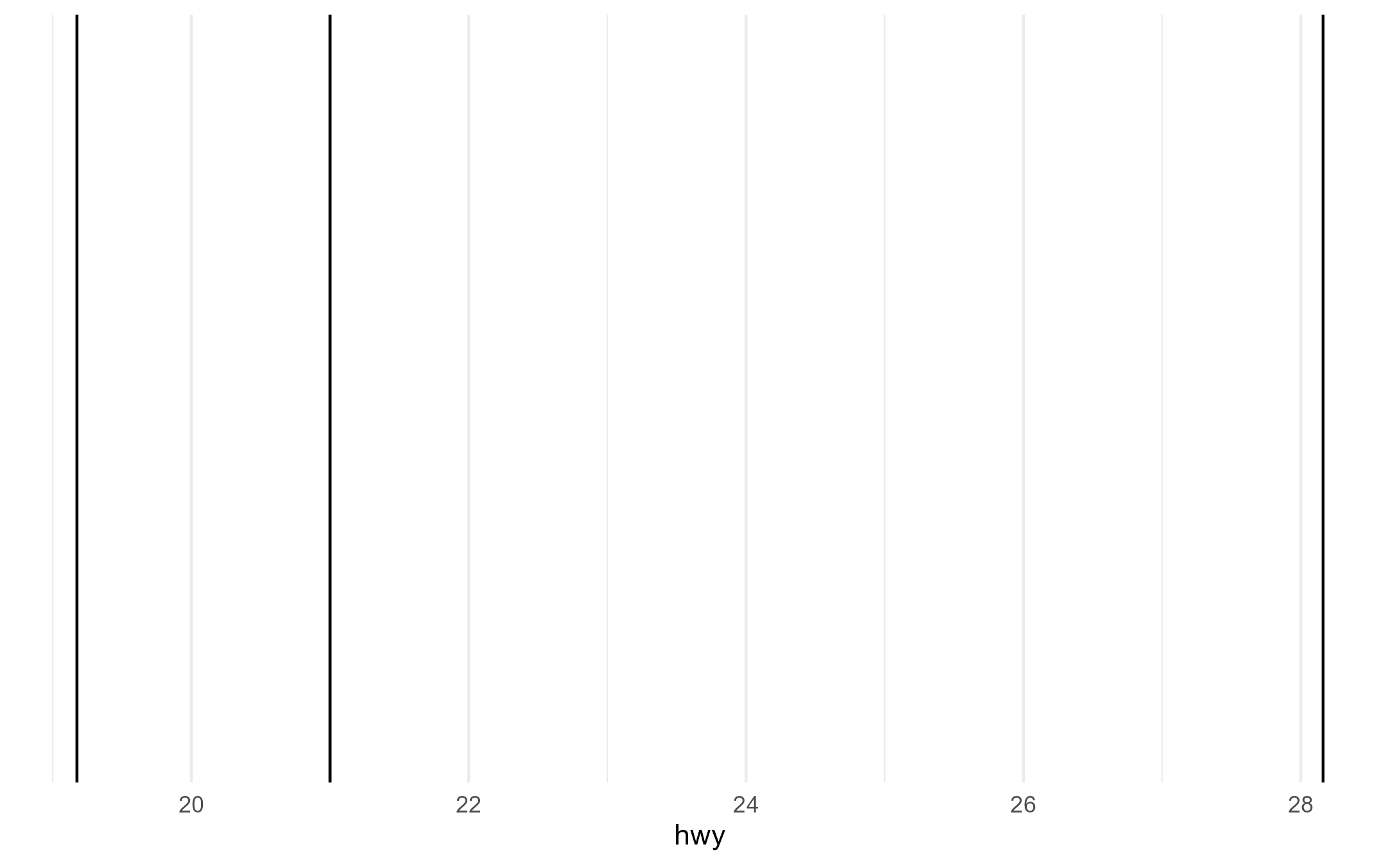
layer_data(p + aes(group = drv))$group
#> [1] 1 2 3Unexpected cases
Here, we’d only like there to be 1 group/line because lines don’t
understand the shape aesthetic
p + aes(shape = drv)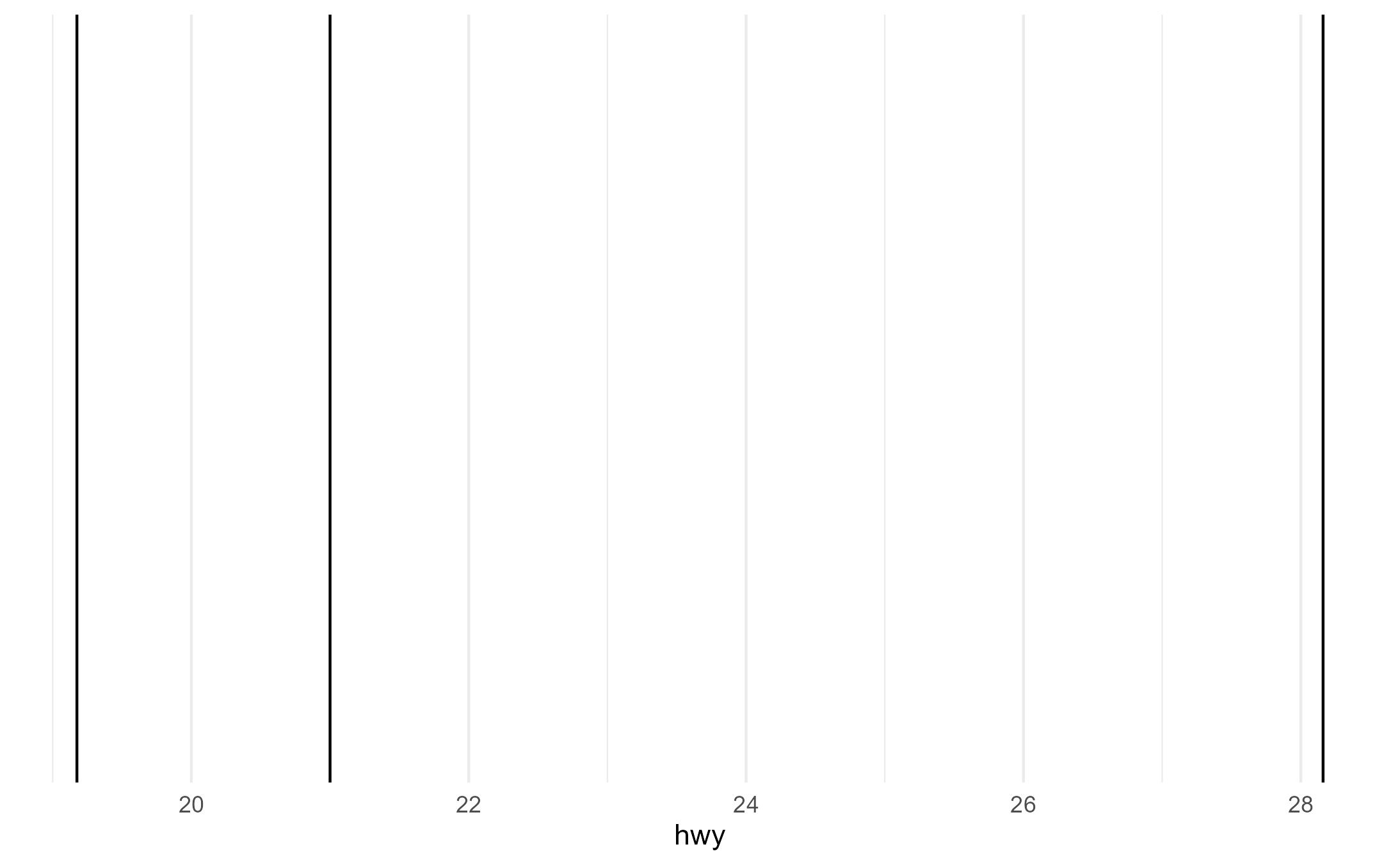
layer_data(p + aes(shape = drv))$group
#> [1] 1 2 3Here, we’d only like there to be 3 groups for drv mapped
to color. Categories in the fl variable should
not interact with drv in the creation of the groups because
lines don’t understand the fill aesthetic.
p + aes(fill = fl, color = drv)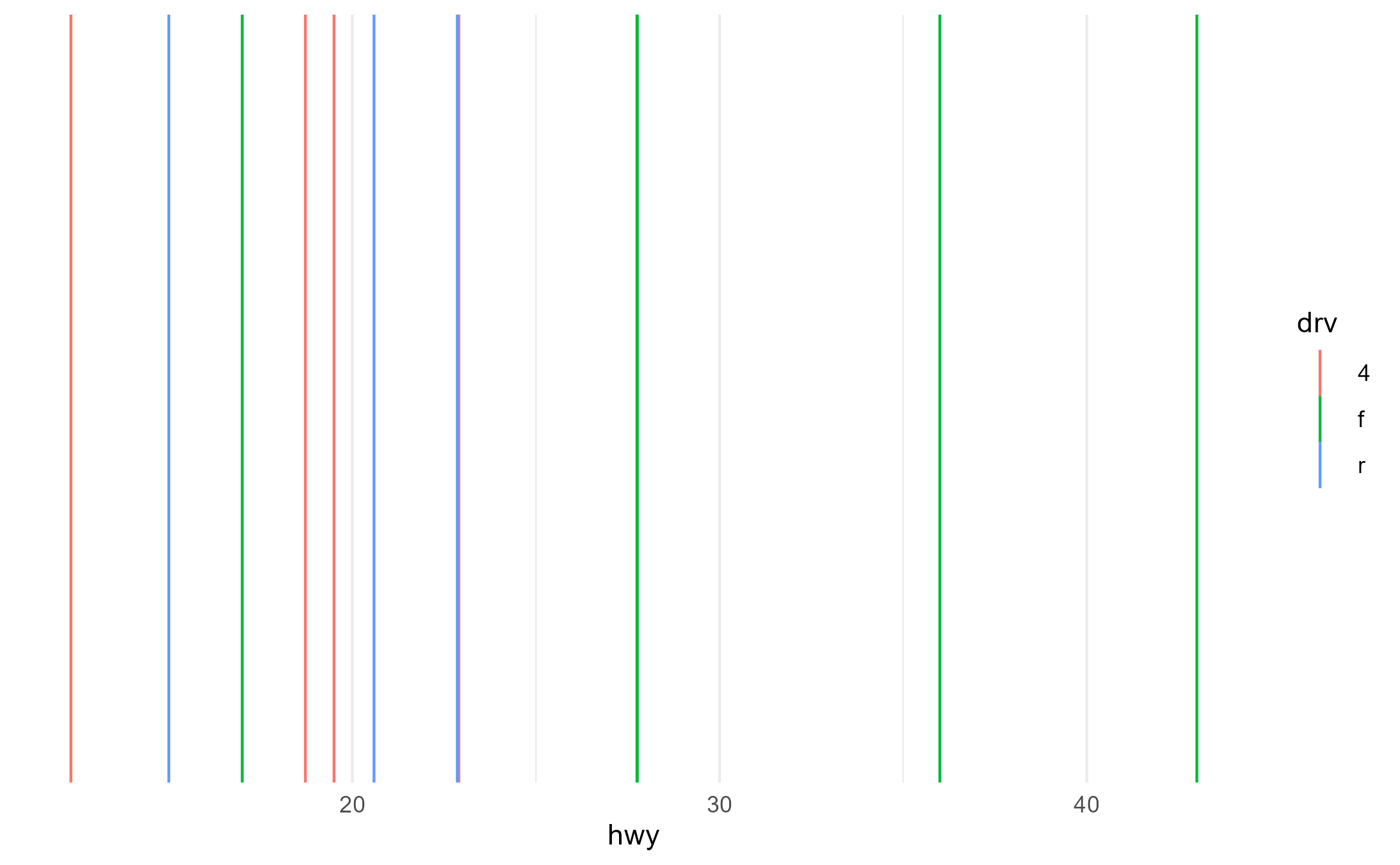
layer_data(p + aes(fill = fl, color = drv))$group
#> [1] 1 2 3 4 5 6 7 8 9 10 11 12Debugging the issue with {ggtrace}
Step 1 - Locating the problem
1.1 Starting at ggplot_build()
As our very first step, let’s see where the groups are being
calculated and added to the data by inspecting
ggplot_build(), the main engine
behind ggplot. For exploratory debugging of ggplot internals,
inspecting the data transformation pipeline in
ggplot_build() is often the best place to start.
If this is your first exposure to ggplot_build(), you
just need to know that its output is basically what’s returned by the
layer_data() function that we demoed above.
layer_data
#> function (plot, i = 1L)
#> {
#> ggplot_build(plot)$data[[i]]
#> }
#> <bytecode: 0x0000020c05f5ffe8>
#> <environment: namespace:ggplot2>We use ggbody() function from {ggtrace} to grab the body
of ggplot_build() as a list, and, with some metaprogramming
with rlang, pick out the steps where the data gets
transformed. Note that the syntax
ggplot2:::ggplot_build.ggplot reflects the fact that
ggplot_build() is an S3 generic defined for the class
<<ggplot>>.
build_pipeline <- ggbody(ggplot2:::ggplot_build.ggplot)
data_assigns <- sapply(build_pipeline,
function(x) {
rlang::is_call(x) &&
!is.null(rlang::call_name(x)) &&
rlang::call_name(x) == "<-" &&
rlang::call_args(x)[[1]] == "data"
}
)
which(data_assigns) # Steps of the build pipeline where data is transformed
#> [1] 8 9 11 12 13 17 18 19 21 22 26 29 30 31You can inspect the source code for ggplot_build() on
the ggplot2
Github repo, and the output of
ggbody(ggplot2:::ggplot_build.ggplot) is also available
below:
Body of ggplot_build()
ggbody(ggplot2:::ggplot_build.ggplot)
#> [[1]]
#> `{`
#>
#> [[2]]
#> plot <- plot_clone(plot)
#>
#> [[3]]
#> if (length(plot$layers) == 0) {
#> plot <- plot + geom_blank()
#> }
#>
#> [[4]]
#> layers <- plot$layers
#>
#> [[5]]
#> layer_data <- lapply(layers, function(y) y$layer_data(plot$data))
#>
#> [[6]]
#> scales <- plot$scales
#>
#> [[7]]
#> by_layer <- function(f) {
#> out <- vector("list", length(data))
#> for (i in seq_along(data)) {
#> out[[i]] <- f(l = layers[[i]], d = data[[i]])
#> }
#> out
#> }
#>
#> [[8]]
#> data <- layer_data
#>
#> [[9]]
#> data <- by_layer(function(l, d) l$setup_layer(d, plot))
#>
#> [[10]]
#> layout <- create_layout(plot$facet, plot$coordinates)
#>
#> [[11]]
#> data <- layout$setup(data, plot$data, plot$plot_env)
#>
#> [[12]]
#> data <- by_layer(function(l, d) l$compute_aesthetics(d, plot))
#>
#> [[13]]
#> data <- lapply(data, scales_transform_df, scales = scales)
#>
#> [[14]]
#> scale_x <- function() scales$get_scales("x")
#>
#> [[15]]
#> scale_y <- function() scales$get_scales("y")
#>
#> [[16]]
#> layout$train_position(data, scale_x(), scale_y())
#>
#> [[17]]
#> data <- layout$map_position(data)
#>
#> [[18]]
#> data <- by_layer(function(l, d) l$compute_statistic(d, layout))
#>
#> [[19]]
#> data <- by_layer(function(l, d) l$map_statistic(d, plot))
#>
#> [[20]]
#> scales_add_missing(plot, c("x", "y"), plot$plot_env)
#>
#> [[21]]
#> data <- by_layer(function(l, d) l$compute_geom_1(d))
#>
#> [[22]]
#> data <- by_layer(function(l, d) l$compute_position(d, layout))
#>
#> [[23]]
#> layout$reset_scales()
#>
#> [[24]]
#> layout$train_position(data, scale_x(), scale_y())
#>
#> [[25]]
#> layout$setup_panel_params()
#>
#> [[26]]
#> data <- layout$map_position(data)
#>
#> [[27]]
#> npscales <- scales$non_position_scales()
#>
#> [[28]]
#> if (npscales$n() > 0) {
#> lapply(data, scales_train_df, scales = npscales)
#> data <- lapply(data, scales_map_df, scales = npscales)
#> }
#>
#> [[29]]
#> data <- by_layer(function(l, d) l$compute_geom_2(d))
#>
#> [[30]]
#> data <- by_layer(function(l, d) l$finish_statistics(d))
#>
#> [[31]]
#> data <- layout$finish_data(data)
#>
#> [[32]]
#> plot$labels$alt <- get_alt_text(plot)
#>
#> [[33]]
#> structure(list(data = data, layout = layout, plot = plot), class = "ggplot_built")After each data transformation step (steps + 1L), let’s
log the data for the first (and only) layer of our reprex plot.
ggtrace(
method = ggplot2:::ggplot_build.ggplot,
trace_steps = which(data_assigns) + 1, # After each data transformation step ...
trace_exprs = quote(data[[1]]), # ... return the data for the first layer
verbose = FALSE
)
#> `ggplot2:::ggplot_build.ggplot` now being traced.
# plot not printed for space
p + aes(fill = drv)
#> Triggering trace on `ggplot2:::ggplot_build.ggplot`
#> Untracing `ggplot2:::ggplot_build.ggplot` on exit.
tracedump1 <- last_ggtrace()Here, tracedump1 is a list of what data
looked like after each data-transformation step inside
ggplot_build(). It’s a list of data frames and there’s a
lot here, but here’s what it looks like after it’s been cleaned up with
tibble::as_tibble()
The evaluated trace expressions captured by last_ggtrace()
lapply(tracedump1, tibble::as_tibble)
#> [[1]]
#> # A tibble: 234 × 11
#> manufacturer model displ year cyl trans drv cty hwy fl class
#> <chr> <chr> <dbl> <int> <int> <chr> <chr> <int> <int> <chr> <chr>
#> 1 audi a4 1.8 1999 4 auto… f 18 29 p comp…
#> 2 audi a4 1.8 1999 4 manu… f 21 29 p comp…
#> 3 audi a4 2 2008 4 manu… f 20 31 p comp…
#> 4 audi a4 2 2008 4 auto… f 21 30 p comp…
#> 5 audi a4 2.8 1999 6 auto… f 16 26 p comp…
#> 6 audi a4 2.8 1999 6 manu… f 18 26 p comp…
#> 7 audi a4 3.1 2008 6 auto… f 18 27 p comp…
#> 8 audi a4 quattro 1.8 1999 4 manu… 4 18 26 p comp…
#> 9 audi a4 quattro 1.8 1999 4 auto… 4 16 25 p comp…
#> 10 audi a4 quattro 2 2008 4 manu… 4 20 28 p comp…
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> [[2]]
#> # A tibble: 234 × 11
#> manufacturer model displ year cyl trans drv cty hwy fl class
#> <chr> <chr> <dbl> <int> <int> <chr> <chr> <int> <int> <chr> <chr>
#> 1 audi a4 1.8 1999 4 auto… f 18 29 p comp…
#> 2 audi a4 1.8 1999 4 manu… f 21 29 p comp…
#> 3 audi a4 2 2008 4 manu… f 20 31 p comp…
#> 4 audi a4 2 2008 4 auto… f 21 30 p comp…
#> 5 audi a4 2.8 1999 6 auto… f 16 26 p comp…
#> 6 audi a4 2.8 1999 6 manu… f 18 26 p comp…
#> 7 audi a4 3.1 2008 6 auto… f 18 27 p comp…
#> 8 audi a4 quattro 1.8 1999 4 manu… 4 18 26 p comp…
#> 9 audi a4 quattro 1.8 1999 4 auto… 4 16 25 p comp…
#> 10 audi a4 quattro 2 2008 4 manu… 4 20 28 p comp…
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> [[3]]
#> # A tibble: 234 × 12
#> manufactu…¹ model displ year cyl trans drv cty hwy fl class PANEL
#> <chr> <chr> <dbl> <int> <int> <chr> <chr> <int> <int> <chr> <chr> <fct>
#> 1 audi a4 1.8 1999 4 auto… f 18 29 p comp… 1
#> 2 audi a4 1.8 1999 4 manu… f 21 29 p comp… 1
#> 3 audi a4 2 2008 4 manu… f 20 31 p comp… 1
#> 4 audi a4 2 2008 4 auto… f 21 30 p comp… 1
#> 5 audi a4 2.8 1999 6 auto… f 16 26 p comp… 1
#> 6 audi a4 2.8 1999 6 manu… f 18 26 p comp… 1
#> 7 audi a4 3.1 2008 6 auto… f 18 27 p comp… 1
#> 8 audi a4 q… 1.8 1999 4 manu… 4 18 26 p comp… 1
#> 9 audi a4 q… 1.8 1999 4 auto… 4 16 25 p comp… 1
#> 10 audi a4 q… 2 2008 4 manu… 4 20 28 p comp… 1
#> # … with 224 more rows, and abbreviated variable name ¹manufacturer
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> [[4]]
#> # A tibble: 234 × 4
#> fill x PANEL group
#> <chr> <int> <fct> <int>
#> 1 f 29 1 2
#> 2 f 29 1 2
#> 3 f 31 1 2
#> 4 f 30 1 2
#> 5 f 26 1 2
#> 6 f 26 1 2
#> 7 f 27 1 2
#> 8 4 26 1 1
#> 9 4 25 1 1
#> 10 4 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> [[5]]
#> # A tibble: 234 × 4
#> fill x PANEL group
#> <chr> <int> <fct> <int>
#> 1 f 29 1 2
#> 2 f 29 1 2
#> 3 f 31 1 2
#> 4 f 30 1 2
#> 5 f 26 1 2
#> 6 f 26 1 2
#> 7 f 27 1 2
#> 8 4 26 1 1
#> 9 4 25 1 1
#> 10 4 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> [[6]]
#> # A tibble: 234 × 4
#> fill x PANEL group
#> <chr> <dbl> <fct> <int>
#> 1 f 29 1 2
#> 2 f 29 1 2
#> 3 f 31 1 2
#> 4 f 30 1 2
#> 5 f 26 1 2
#> 6 f 26 1 2
#> 7 f 27 1 2
#> 8 4 26 1 1
#> 9 4 25 1 1
#> 10 4 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> [[7]]
#> # A tibble: 3 × 4
#> x fill PANEL group
#> <dbl> <chr> <fct> <int>
#> 1 19.2 4 1 1
#> 2 28.2 f 1 2
#> 3 21 r 1 3
#>
#> [[8]]
#> # A tibble: 3 × 4
#> x fill PANEL group
#> <dbl> <chr> <fct> <int>
#> 1 19.2 4 1 1
#> 2 28.2 f 1 2
#> 3 21 r 1 3
#>
#> [[9]]
#> # A tibble: 3 × 4
#> x fill PANEL group
#> <dbl> <chr> <fct> <int>
#> 1 19.2 4 1 1
#> 2 28.2 f 1 2
#> 3 21 r 1 3
#>
#> [[10]]
#> # A tibble: 3 × 4
#> x fill PANEL group
#> <dbl> <chr> <fct> <int>
#> 1 19.2 4 1 1
#> 2 28.2 f 1 2
#> 3 21 r 1 3
#>
#> [[11]]
#> # A tibble: 3 × 4
#> x fill PANEL group
#> <dbl> <chr> <fct> <int>
#> 1 19.2 4 1 1
#> 2 28.2 f 1 2
#> 3 21 r 1 3
#>
#> [[12]]
#> # A tibble: 3 × 8
#> fill x PANEL group colour size linetype alpha
#> <chr> <dbl> <fct> <int> <chr> <dbl> <dbl> <lgl>
#> 1 #F8766D 19.2 1 1 black 0.5 1 NA
#> 2 #00BA38 28.2 1 2 black 0.5 1 NA
#> 3 #619CFF 21 1 3 black 0.5 1 NA
#>
#> [[13]]
#> # A tibble: 3 × 8
#> fill x PANEL group colour size linetype alpha
#> <chr> <dbl> <fct> <int> <chr> <dbl> <dbl> <lgl>
#> 1 #F8766D 19.2 1 1 black 0.5 1 NA
#> 2 #00BA38 28.2 1 2 black 0.5 1 NA
#> 3 #619CFF 21 1 3 black 0.5 1 NA
#>
#> [[14]]
#> # A tibble: 3 × 8
#> fill x PANEL group colour size linetype alpha
#> <chr> <dbl> <fct> <int> <chr> <dbl> <dbl> <lgl>
#> 1 #F8766D 19.2 1 1 black 0.5 1 NA
#> 2 #00BA38 28.2 1 2 black 0.5 1 NA
#> 3 #619CFF 21 1 3 black 0.5 1 NAWe can now map over every element of the list to see where the
group column was first present.
group_present1 <- sapply(tracedump1, function(x) {
"group" %in% colnames(x) && sort(unique(x$group)) == 1:3
})
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
group_present1
#> [1] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
#> [13] TRUE TRUEStep 12 of ggplot_build() is where group
first appears. We suspect that that’s where group is
computed and assigned
which(data_assigns)[min(which(group_present1))]
#> [1] 12
ggbody(ggplot2:::ggplot_build.ggplot)[[12]]
#> data <- by_layer(function(l, d) l$compute_aesthetics(d, plot))The code from Step 12 of ggplot_build() calls the
compute_aesthetics method of each layer l. The
by_layer() function defined in Step 7 of
ggplot_build() is what’s doing this.
The internal function by_layer()
ggbody(ggplot2:::ggplot_build.ggplot)[[7]]
#> by_layer <- function(f) {
#> out <- vector("list", length(data))
#> for (i in seq_along(data)) {
#> out[[i]] <- f(l = layers[[i]], d = data[[i]])
#> }
#> out
#> }1.2 Digging deeper into Layer$compute_aesthetics()
At this point we realize that we have to go a little deeper than
ggplot_build(). We need to inspect
Layer$compute_aesthetics - the
compute_aesthetics method of the Layer
ggproto.
(Check out the quick aside on the Layer ggproto below if
this is your first time encountering it, but for the purposes of this
vignette you just need to know that
Layer$compute_aesthetics is a function called inside
ggplot_build() where we think group is being
calculated.)
An aside on the Layer ggproto
A Layer is an object returned by
ggplot2::layer(). You don’t see that function often, but
it’s what gets returned by geom_*() and
stat_*() functions, so you’ve actually seen this a lot.
For example, if you call geom_bar() on its own, you’ll
get back an object of class
LayerInstance/Layer
geom_bar()
#> geom_bar: width = NULL, na.rm = FALSE, orientation = NA
#> stat_count: width = NULL, na.rm = FALSE, orientation = NA
#> position_stack
class(geom_bar())
#> [1] "LayerInstance" "Layer" "ggproto" "gg"The body of geom_bar() just calls layer()
with some defaults optimized for drawing bar plots.
body(geom_bar)
#> {
#> layer(data = data, mapping = mapping, stat = stat, geom = GeomBar,
#> position = position, show.legend = show.legend, inherit.aes = inherit.aes,
#> params = list(width = width, na.rm = na.rm, orientation = orientation,
#> ...))
#> }So these two are practically the same:
geom_bar()
#> geom_bar: width = NULL, na.rm = FALSE, orientation = NA
#> stat_count: width = NULL, na.rm = FALSE, orientation = NA
#> position_stack
layer(
geom = GeomBar, stat = StatCount, position = PositionStack,
params = list(width = NULL, na.rm = FALSE, orientation = NA)
)
#> geom_bar: width = NULL, na.rm = FALSE, orientation = NA
#> stat_count: width = NULL, na.rm = FALSE, orientation = NA
#> position_stackThese Layer objects have various properties and methods,
including compute_aesthetics.
names(as.list(geom_bar()))
#> [1] "mapping" "geom_params" "layer_data"
#> [4] "compute_statistic" "computed_mapping" "compute_aesthetics"
#> [7] "stat_params" "map_statistic" "draw_geom"
#> [10] "stat" "setup_layer" "inherit.aes"
#> [13] "finish_statistics" "geom" "compute_position"
#> [16] "position" "print" "data"
#> [19] "aes_params" "computed_stat_params" "computed_geom_params"
#> [22] "compute_geom_1" "compute_geom_2" "show.legend"The compute_aesthetics method is defined in the parent
Layer ggproto which is unexported. The layer returned by
geom_bar() simply inherits this method, and this is
reflected in the fact that it has the class LayerInstance,
as we also so above.
class(ggplot2:::Layer)
#> [1] "Layer" "ggproto" "gg"
class(geom_bar())
#> [1] "LayerInstance" "Layer" "ggproto" "gg"
ggbody(ggplot2:::Layer$compute_aesthetics)
#> [[1]]
#> `{`
#>
#> [[2]]
#> aesthetics <- self$computed_mapping
#>
#> [[3]]
#> set <- names(aesthetics) %in% names(self$aes_params)
#>
#> [[4]]
#> calculated <- is_calculated_aes(aesthetics)
#>
#> [[5]]
#> modifiers <- is_scaled_aes(aesthetics)
#>
#> [[6]]
#> aesthetics <- aesthetics[!set & !calculated & !modifiers]
#>
#> [[7]]
#> if (!is.null(self$geom_params$group)) {
#> aesthetics[["group"]] <- self$aes_params$group
#> }
#>
#> [[8]]
#> scales_add_defaults(plot$scales, data, aesthetics, plot$plot_env)
#>
#> [[9]]
#> env <- child_env(baseenv(), stage = stage)
#>
#> [[10]]
#> evaled <- lapply(aesthetics, eval_tidy, data = data, env = env)
#>
#> [[11]]
#> evaled <- compact(evaled)
#>
#> [[12]]
#> warn_for_aes_extract_usage(aesthetics, data[setdiff(names(data),
#> "PANEL")])
#>
#> [[13]]
#> nondata_cols <- check_nondata_cols(evaled)
#>
#> [[14]]
#> if (length(nondata_cols) > 0) {
#> msg <- paste0("Aesthetics must be valid data columns. Problematic aesthetic(s): ",
#> paste0(vapply(nondata_cols, function(x) {
#> paste0(x, " = ", as_label(aesthetics[[x]]))
#> }, character(1)), collapse = ", "), ". \nDid you mistype the name of a data column or forget to add after_stat()?")
#> abort(msg)
#> }
#>
#> [[15]]
#> n <- nrow(data)
#>
#> [[16]]
#> if (n == 0) {
#> if (length(evaled) == 0) {
#> n <- 0
#> }
#> else {
#> n <- max(vapply(evaled, length, integer(1)))
#> }
#> }
#>
#> [[17]]
#> check_aesthetics(evaled, n)
#>
#> [[18]]
#> if (empty(data) && n > 0) {
#> evaled$PANEL <- 1
#> } else {
#> evaled$PANEL <- data$PANEL
#> }
#>
#> [[19]]
#> evaled <- lapply(evaled, unname)
#>
#> [[20]]
#> evaled <- as_gg_data_frame(evaled)
#>
#> [[21]]
#> evaled <- add_group(evaled)
#>
#> [[22]]
#> evaledLet’s now repeat the same know-nothing, brute-force exploratory
process inside Layer$compute_aesthetics. Here, we inspect
the output of every line using the ~step keyword inside the
trace_exprs argument to locate where group get
computed.
ggtrace(
method = ggplot2:::Layer$compute_aesthetics,
trace_steps = seq_along(ggbody(ggplot2:::Layer$compute_aesthetics)),
trace_exprs = quote(~step),
verbose = FALSE
)
#> `ggplot2:::Layer$compute_aesthetics` now being traced.
# plot not printed for space
p + aes(fill = drv)
#> Triggering trace on `ggplot2:::Layer$compute_aesthetics`
#> Untracing `ggplot2:::Layer$compute_aesthetics` on exit.
tracedump2 <- last_ggtrace()
group_present2 <- sapply(tracedump2, function(x) {
"group" %in% colnames(x) && sort(unique(x$group)) == 1:3
})
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
#> Warning in "group" %in% colnames(x) && sort(unique(x$group)) == 1:3: 'length(x)
#> = 3 > 1' in coercion to 'logical(1)'
length(group_present2)
#> [1] 22
which(group_present2)
#> [1] 21 22Step 21 of Layer$compute_aesthetics seems to be where
this happens. Here, it calls an unexported internal utility function
called add_group(), and if we look at the source
code we can confirm that we have indeed located where
group is calculated and assigned.
min(which(group_present2))
#> [1] 21
ggbody(ggplot2:::Layer$compute_aesthetics)[[21]]
#> evaled <- add_group(evaled)But that only gets us half-way there to solving the issue because we
can’t/shouldn’t modify the Layer ggproto in extension packages - it’s
unexported! Plus, we want to change the behavior for
geom_x_mean() specifically, such that the aesthetics that
it can’t understand are ignored for the calculation of its own
groups, but not for the groups of other layers.
Step 2 - Identifying the extension point
At this point, we have to hunt down the next time that the
Layer dispatches a Stat or Geom ggproto method with the transformed data
after group is calculated, so that we can undo the grouping
calculation inside Stat/Geom as necessary (since Stat and
Geom are exported and are the recommended extension
points).
Going back to ggplot_build(), the next
l$<method> call after
l$compute_aesthetics is the compute_statistic
method at Step 18
ggbody(ggplot2:::ggplot_build.ggplot)[[18]]
#> data <- by_layer(function(l, d) l$compute_statistic(d, layout))Let’s see what happens inside the
Layer$compute_statistic method
ggbody(ggplot2:::Layer$compute_statistic)
#> [[1]]
#> `{`
#>
#> [[2]]
#> if (empty(data)) return(new_data_frame())
#>
#> [[3]]
#> self$computed_stat_params <- self$stat$setup_params(data, self$stat_params)
#>
#> [[4]]
#> data <- self$stat$setup_data(data, self$computed_stat_params)
#>
#> [[5]]
#> self$stat$compute_layer(data, self$computed_stat_params, layout)We see that the next time the data gets transformed after being
passed to Layer$compute_statistic is by the
setup_data method of the layer’s Stat ggproto at Step
4.
ggbody(ggplot2:::Layer$compute_statistic)[[4]]
#> data <- self$stat$setup_data(data, self$computed_stat_params)Let’s see what kind of information we have at this stage, and whether
the information is sufficient for us to implement our desired design
change to geom_x_mean()
Step 2.1 - Inspecting the setup_data method
First, let’s figure out what the Stat ggproto associated with
geom_x_mean() is
class(geom_x_mean()$stat)[1]
#> [1] "StatXmean"And let’s inspect its setup_data method
ggbody(ggxmean:::StatXmean$setup_data) # errors!
#> Error:
#> ! Method 'setup_data' is not defined for `ggxmean:::StatXmean`
#> Check inheritance with `get_method_inheritance(ggxmean:::StatXmean)`It looks like setup_data is inherited from a parent
ggproto. The only candidate here from looking at its
class() is the Stat parent ggproto.
class(ggxmean:::StatXmean)
#> [1] "StatXmean" "Stat" "ggproto" "gg"If we decide to change the behavior of setup_data for
StatXmean, we’ll need to define its own version of the method, instead
of having it simlpy inherit Stat$setup_data.
For now, let’s grab the inherited method from Stat and proceed with
inspecting its behavior. With the inherit = TRUE argument
inside ggbody(), we confirm that StatXmean
indeed inherits Stat$setup_data
ggbody(ggxmean:::StatXmean$setup_data, inherit = TRUE)
#> Method inherited from `Stat$setup_data`
#> [[1]]
#> `{`
#>
#> [[2]]
#> dataOkay so this method just returns the data by default. But what does
data look like in practice?
Step 2.2 - Logging the execution of
Stat$setup_data
The following ggtrace() code is a shorthand for
inserting a trace that simply returns the output of the last step
(-1) of the method:
ggtrace(Stat$setup_data, -1, verbose = FALSE)
#> `Stat$setup_data` now being traced.
# plot not printed for space
p + aes(fill = drv)
#> Triggering trace on `Stat$setup_data`
#> Untracing `Stat$setup_data` on exit.We see that the data contains information about the supplied
aesthetics and the calculated PANEL and group
information.
tibble::as_tibble(last_ggtrace()[[1]])
#> # A tibble: 234 × 4
#> fill x PANEL group
#> <chr> <dbl> <fct> <int>
#> 1 f 29 1 2
#> 2 f 29 1 2
#> 3 f 31 1 2
#> 4 f 30 1 2
#> 5 f 26 1 2
#> 6 f 26 1 2
#> 7 f 27 1 2
#> 8 4 26 1 1
#> 9 4 25 1 1
#> 10 4 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rowsThis is sufficient information for our purposes.
Step 3 - Developing a solution
Step 3.1 - Devising a plan (pseudo-code outline)
Here’s the plan. Instead of having the setup_data method
just return the data it receives, let’s have it check for two
conditions:
- Is the group explicitly supplied?
- If so, leave the data alone. Otherwise…
- Are all the discrete variable used to calculate groups
understandable by the geom?
- If so, leave the data alone. Otherwise, re-calculate groups by
dropping those extraneous variables and passing the data through
ggplot2:::add_group()again.
- If so, leave the data alone. Otherwise, re-calculate groups by
dropping those extraneous variables and passing the data through
Step 3.2 - Setting up for testing
One of the biggest perks of {ggtrace} is the ability to log the
output of the triggered traces. We’ve been seeing a bit of that with
last_ggtrace() but we can scale this up.
Using global_ggtrace() and persistent tracing with
ggtrace(once = FALSE), we can capture what
data looks like inside the setup_data method
across our two expected and two unexpected cases from our reprex.
# Turn the global tracedump on
global_ggtrace_on()
#> Global tracedump activated.
ggtrace(Stat$setup_data, -1, once = FALSE, verbose = FALSE)
#> `Stat$setup_data` now being traced.
#> Creating a persistent trace. Remember to `gguntrace(Stat$setup_data)`!
# plots not printed for space
p + aes(color = drv)
#> Triggering persistent trace on `Stat$setup_data`
p + aes(group = drv)
#> Triggering persistent trace on `Stat$setup_data`
p + aes(shape = drv)
#> Triggering persistent trace on `Stat$setup_data`
p + aes(fill = fl, color = drv)
#> Triggering persistent trace on `Stat$setup_data`
tracedump3 <- global_ggtrace()
tracedump3 <- lapply(tracedump3, `[[`, 1) # simplify
# Clean up and turn the global tracedump back off
gguntrace(Stat$setup_data)
#> `Stat$setup_data` no longer being traced.
clear_global_ggtrace()
#> Global tracedump cleared.
global_ggtrace_off()
#> Global tracedump deactivated.
# Label the trace dumps
names(tracedump3) <- c(paste0("expected", 1:2), paste0("unexpected", 1:2))
names(tracedump3)
#> [1] "expected1" "expected2" "unexpected1" "unexpected2"Now tracedump3 holds the value for data
from inside the setup_data method for our four plots of
interest.
The value of tracedump3
lapply(tracedump3, tibble::as_tibble)
#> $expected1
#> # A tibble: 234 × 4
#> colour x PANEL group
#> <chr> <dbl> <fct> <int>
#> 1 f 29 1 2
#> 2 f 29 1 2
#> 3 f 31 1 2
#> 4 f 30 1 2
#> 5 f 26 1 2
#> 6 f 26 1 2
#> 7 f 27 1 2
#> 8 4 26 1 1
#> 9 4 25 1 1
#> 10 4 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> $expected2
#> # A tibble: 234 × 3
#> x group PANEL
#> <dbl> <int> <fct>
#> 1 29 2 1
#> 2 29 2 1
#> 3 31 2 1
#> 4 30 2 1
#> 5 26 2 1
#> 6 26 2 1
#> 7 27 2 1
#> 8 26 1 1
#> 9 25 1 1
#> 10 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> $unexpected1
#> # A tibble: 234 × 4
#> shape x PANEL group
#> <chr> <dbl> <fct> <int>
#> 1 f 29 1 2
#> 2 f 29 1 2
#> 3 f 31 1 2
#> 4 f 30 1 2
#> 5 f 26 1 2
#> 6 f 26 1 2
#> 7 f 27 1 2
#> 8 4 26 1 1
#> 9 4 25 1 1
#> 10 4 28 1 1
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rows
#>
#> $unexpected2
#> # A tibble: 234 × 5
#> fill colour x PANEL group
#> <chr> <chr> <dbl> <fct> <int>
#> 1 p f 29 1 8
#> 2 p f 29 1 8
#> 3 p f 31 1 8
#> 4 p f 30 1 8
#> 5 p f 26 1 8
#> 6 p f 26 1 8
#> 7 p f 27 1 8
#> 8 p 4 26 1 7
#> 9 p 4 25 1 7
#> 10 p 4 28 1 7
#> # … with 224 more rows
#> # ℹ Use `print(n = ...)` to see more rowsStep 3.3 - Tackling the problem piece by piece
Part 1 - Check if group is derived
Our first condition checking for whether group is
explicitly supplied is actually very easy to implement.
Going back to Layer$compute_aesthetics, we can see that
the PANEL column is assigned first and then the
group column is assigned afterwards. This means that if
group did not already exist, it’d appear to the
right of PANEL. But if it did exist before, it’d
just be modified in its original position and appear to the
left of PANEL (since PANEL can never
exist before).
We can confirm this from our two expected cases:
colnames(tracedump3$expected1) # `aes(color = drv)`
#> [1] "colour" "x" "PANEL" "group"
colnames(tracedump3$expected2) # `aes(group = drv)`
#> [1] "x" "group" "PANEL"We can make a function that checks for the position of the
group column relative to the PANEL column, and
use our tracedump to validate it
group_is_derived <- function(data) {
# Do "group" and "PANEL" appear out of order?
is.unsorted(match(c("group", "PANEL"), colnames(data)))
}
group_is_derived(tracedump3$expected1)
#> [1] TRUE
group_is_derived(tracedump3$expected2)
#> [1] FALSEPart 2 - Check if the geom can understand all discrete aesthetics
For our second condition, we need to figure out which aesthetics our geom can understand.
First, we check what geom geom_x_mean() uses. We see
that it uses the unexported geom ggxmean:::GeomXline.
class(geom_x_mean()$geom)[1]
#> [1] "GeomXline"Then we gather all the aesthetics that it can handle
understandable_aes <- unique(c(
ggxmean:::GeomXline$required_aes,
names(ggxmean:::GeomXline$default_aes),
ggxmean:::GeomXline$optional_aes
))
understandable_aes
#> [1] "x" "colour" "size" "linetype" "alpha"We just want to check among the non-positional aesthetics, so this is the set we want:
We can now write a function that returns the discrete variables in the data that cannot be understood by GeomXline.
not_understandable_aes <- function(data) {
discretes <- colnames(data)[sapply(data, ggplot2:::is.discrete)]
discretes <- discretes[!discretes %in% c("group", "PANEL")]
discretes[!discretes %in% c("colour", "size", "linetype", "alpha")]
}We then validate the function against our tracedump
Part 3 - Handle the exception
Lastly, in the case where groups were computed using discrete variables mapped to aesthetics that GeomXline doesn’t understand, we want to re-assign groups.
The following function takes the data, strips it of the offending variables/columns, and returns a vector of the new groupings.
retrained_groups <- function(data) {
not_understandables <- not_understandable_aes(data)
data <- data[!colnames(data) %in% c("group", not_understandables)]
ggplot2:::add_group(data)$group
}This returns us the expected number of groups from our unexpected
reprex cases (recall that unexpected2 used to have 12
groups but now has 3 because it’s correctly ignoring the interaction
with fill = fl).
Part 4 - Bring it together
Now we melt the functions from Parts 1-3 into a single function that
will become our new setup_data method for
StatXmean.
setup_data_new <- function(data) {
if (is.unsorted(match(c("group", "PANEL"), colnames(data)))) {
discretes <- colnames(data)[sapply(data, ggplot2:::is.discrete)]
discretes <- discretes[!discretes %in% c("group", "PANEL")]
not_understandables <- discretes[!discretes %in% c("colour", "size", "linetype", "alpha")]
if (length(not_understandables) > 0) {
dummy_data <- data[!colnames(data) %in% c("group", not_understandables)]
data$group <- ggplot2:::add_group(dummy_data)$group
}
}
data
}We again validate it against our expected and unexpected cases and see that it indeed works!
tracedump3_modified <- lapply(tracedump3, setup_data_new)
lapply(tracedump3_modified, function(x) sort(unique(x$group) ))
#> $expected1
#> [1] 1 2 3
#>
#> $expected2
#> [1] 1 2 3
#>
#> $unexpected1
#> [1] -1
#>
#> $unexpected2
#> [1] 1 2 3*Note that we’re still using unexported ggplot2 functions here like
is.discrete and add_group. You’d normally
refactor them or copy them over before releasing the fix, but we’ll skip
that here for the sake of time.
Step 3.4 - Implementing the solution
We’d like to test this whole solution out as a user-facing layer before we call it a day.
There are just two important pieces here:
We create a ggproto class called StatXmean2 which is an exact copy of StatXmean except it also defines its own
setup_datamethod as we defined above.We create a layer called
geom_x_mean2()which is an exact copy ofgeom_x_mean()except that thestatargument ofggplot2::layer()is StatXmean2
StatXmean2 <- ggplot2::ggproto(
"StatXmean2",
ggplot2::Stat,
setup_data = function(data, params) {
if (is.unsorted(match(c("group", "PANEL"), colnames(data)))) {
discretes <- colnames(data)[sapply(data, ggplot2:::is.discrete)]
discretes <- discretes[!discretes %in% c("group", "PANEL")]
not_understandables <- discretes[!discretes %in% c("colour", "size", "linetype", "alpha")]
if (length(not_understandables) > 0) {
dummy_data <- data[!colnames(data) %in% c("group", not_understandables)]
data$group <- ggplot2:::add_group(dummy_data)$group
}
}
data
},
compute_group = function(data, scales) {
data.frame(x = mean(data$x))
},
required_aes = c("x")
)
geom_x_mean2 <- function(mapping = NULL, data = NULL,
position = "identity", na.rm = FALSE, show.legend = NA,
inherit.aes = TRUE, ...) {
ggplot2::layer(
stat = StatXmean2, geom = ggxmean:::GeomXline, data = data, mapping = mapping,
position = position, show.legend = show.legend, inherit.aes = inherit.aes,
params = list(na.rm = na.rm, ...)
)
}Our final check against our original reprexes shows that it works as intended
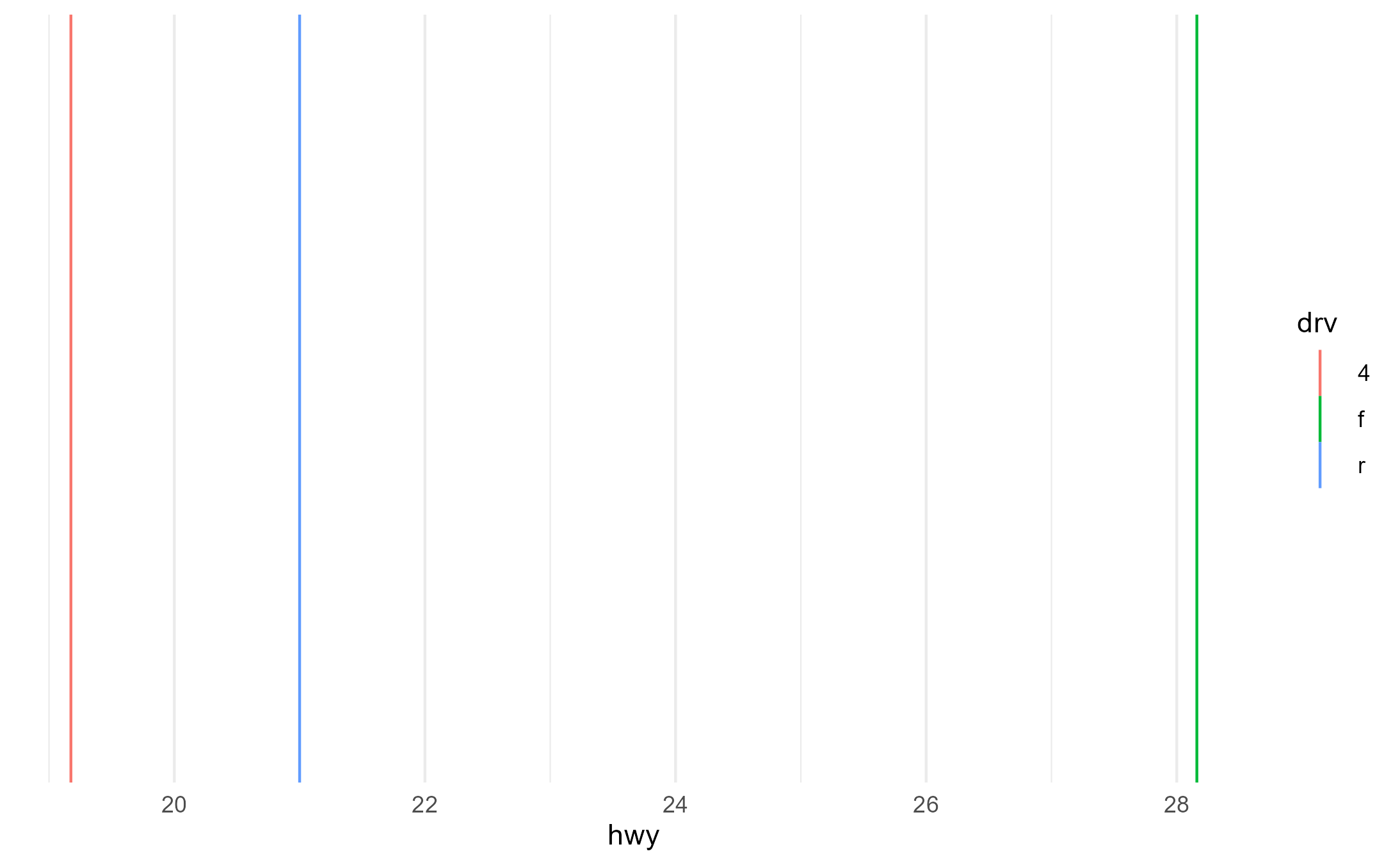
p2 + aes(group = drv)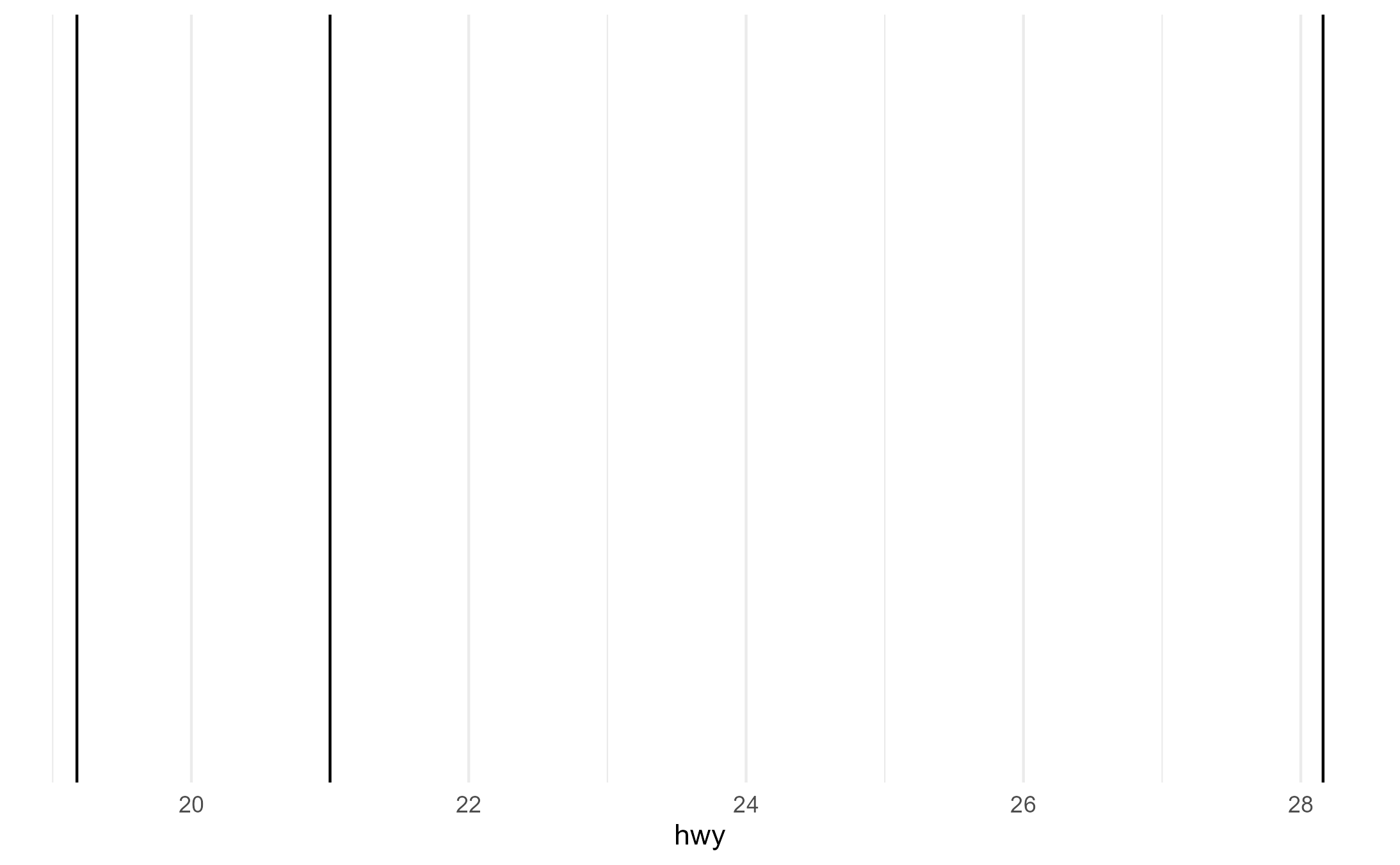
p2 + aes(shape = drv)
p2 + aes(fill = fl, color = drv)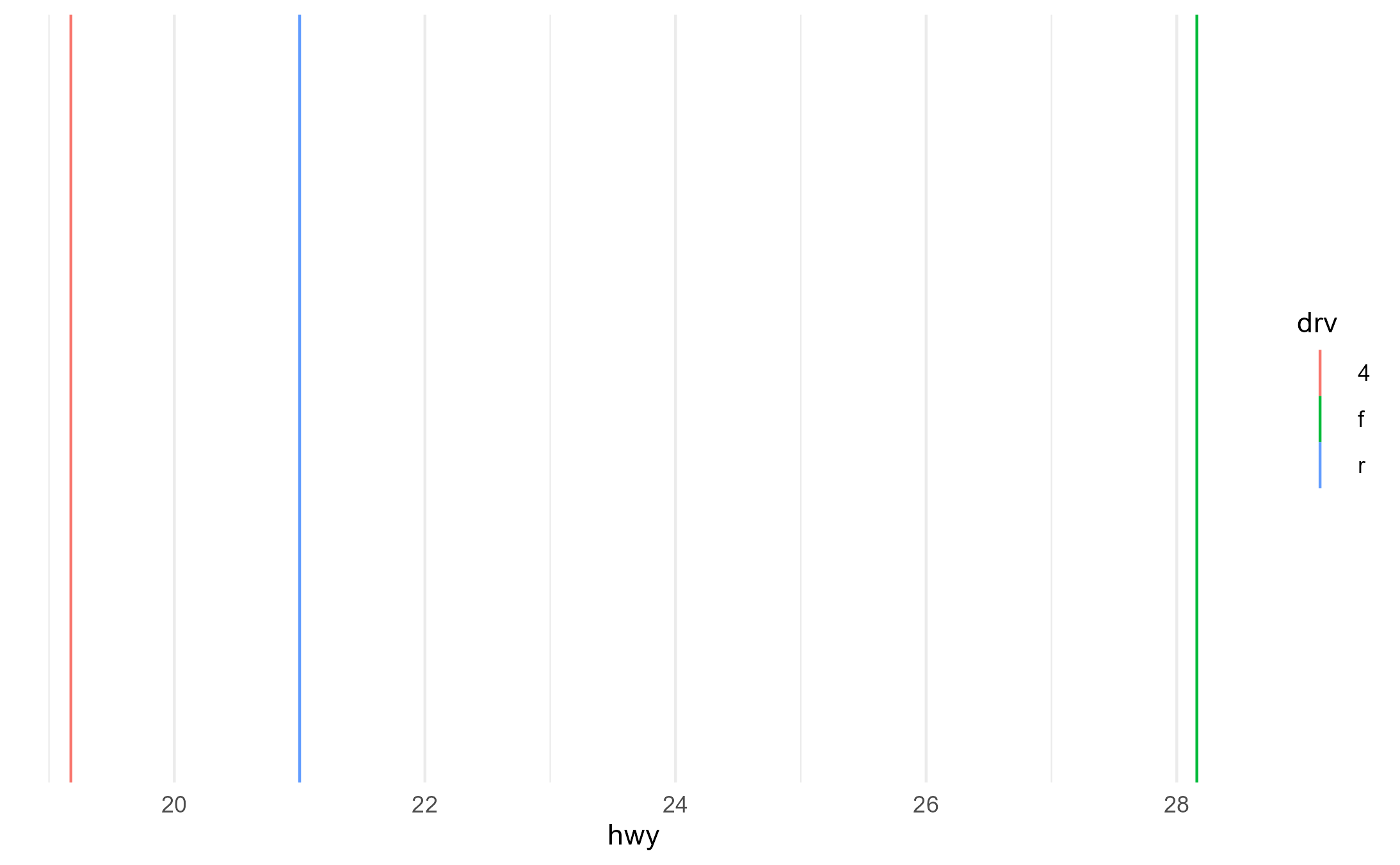
Step 3.5 - Testing the solution
If you want to write tests, we can also check whether solution generalizes to other cases.
Here, we can see it correctly works with facets even when some groups are missing in a facet, so that’s good
p2 +
aes(color = drv) +
facet_wrap(~fl)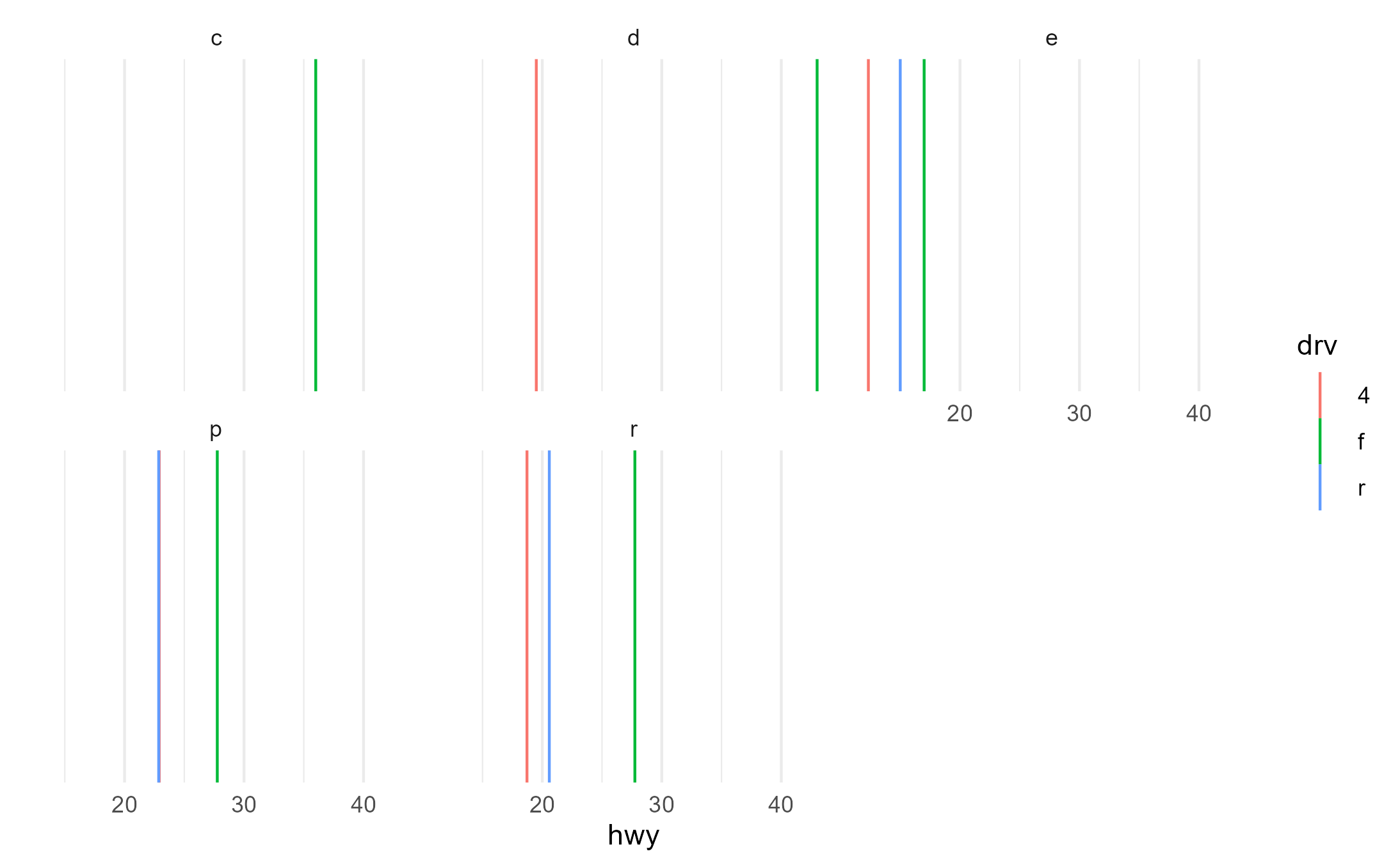
And we can also see that it fails the way it should when it’s
supplied an explicit group aesthetic but also receives another discrete
mapping to color
p2 + aes(group = fl, color = drv)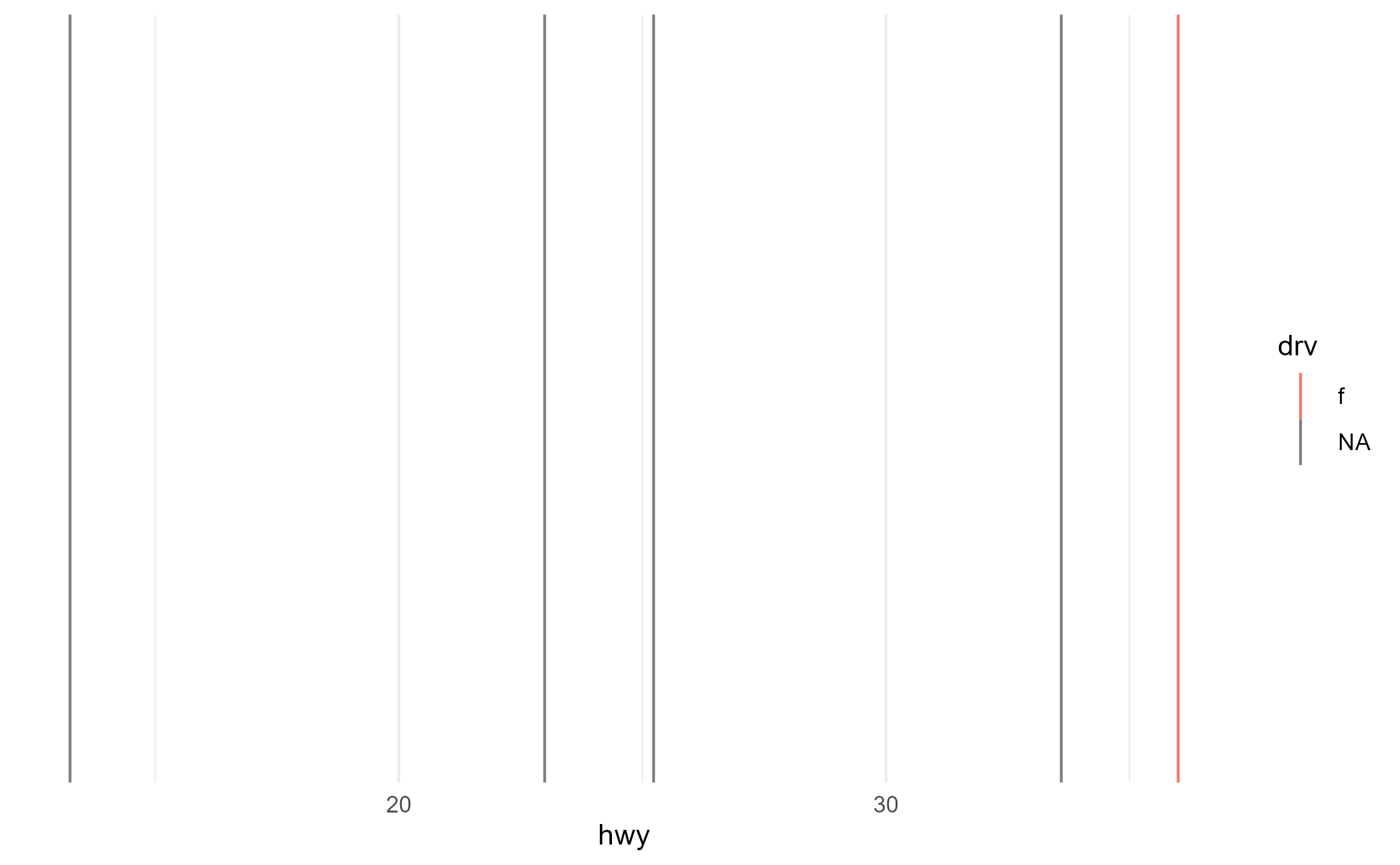
This is because there’s one unique match between
fl == "c" and drv== "f", but no unique value
for drv for the other four fl categories. So
the color scale assigns a color to one of the lines but then for the
other four lines it just gives up and returns NAs (which are colored
grey by default)
table(mpg$drv, mpg$fl)
#>
#> c d e p r
#> 4 0 2 6 20 75
#> f 1 3 1 25 76
#> r 0 0 1 7 17
p2 +
aes(group = fl, color = drv) +
geom_label(aes(label = fl, y = 0), stat = StatXmean2)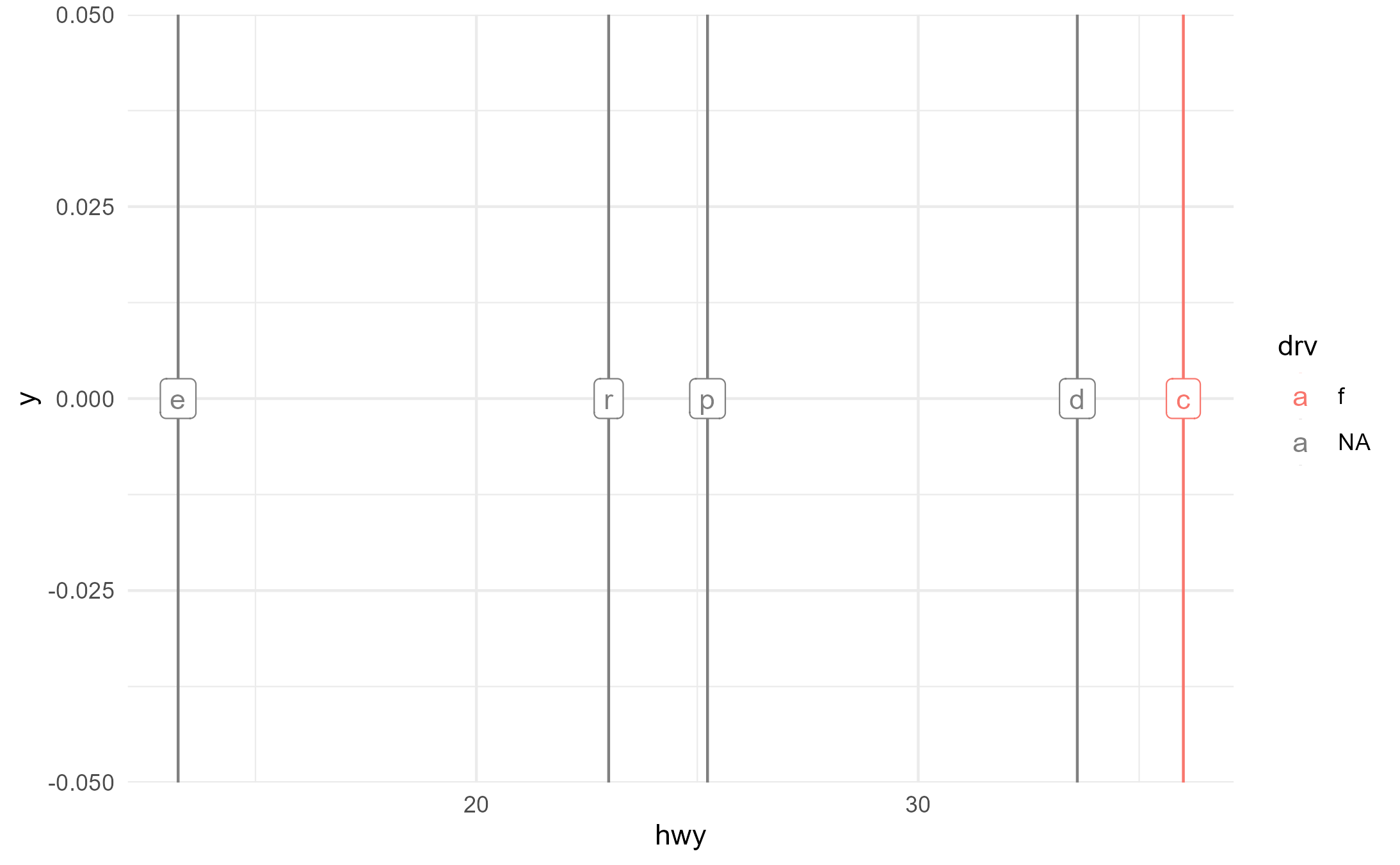
Wrapping up
We’re satisfied so we can now turn development mode off, which lets us access our current installation of {ggxmean} again. We can now also test our solution against the current version of the package to see whether it’d still work (although we skip that step here).
devtools::dev_mode(on = FALSE)
#> ✔ Dev mode: OFF
devtools::package_info("ggxmean", FALSE, FALSE)$source
#> [1] "local"Finally, since this vignette covered a real-world debugging scenario, the solution has been submitted as a PR!
Many thanks to Gina for allowing me to use {ggxmean} as a case study for exploratory debugging with {ggtrace}!
